DML ATE Example#
This notebook covers scenario:
Is RCT |
Treatment |
Outcome |
EDA |
Estimands |
Refutation |
|---|---|---|---|---|---|
Observational |
Binary |
Continuous |
Yes |
ATE |
Yes |
We will estimate Average Treatment Effect (ATE) of binary treatment on continuous outcome. It shows explonatary data analysis and refutation tests
Generate data#
Let’s generate data of how feature (Treatment) impact on ARPU (Outcome) with linear effect (theta) = 1.8
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from causalkit.data import CausalDatasetGenerator, CausalData
# Reproducibility
np.random.seed(42)
confounder_specs = [
{"name": "tenure_months", "dist": "normal", "mu": 24, "sd": 12},
{"name": "avg_sessions_week", "dist": "normal", "mu": 5, "sd": 2},
{"name": "spend_last_month", "dist": "uniform", "a": 0, "b": 200},
{"name": "premium_user", "dist": "bernoulli", "p": 0.25},
{"name": "urban_resident", "dist": "bernoulli", "p": 0.60},
]
# Causal effect and noise
theta = 1.8 # ATE: +1.8 ARPU units if new_feature = 1
sigma_y = 3.5 # ARPU noise std
target_t_rate = 0.35 # ~35% treated
# Effects of confounders on ARPU (baseline, additive)
# Order: tenure_months, avg_sessions_week, spend_last_month, premium_user, urban_resident
beta_y = np.array([
0.05, # tenure_months: small positive effect
0.40, # avg_sessions_week: strong positive effect
0.02, # spend_last_month: recent spend correlates with ARPU
2.00, # premium_user: premium users have higher ARPU
1.00, # urban_resident: urban users slightly higher ARPU
], dtype=float)
# Effects of confounders on treatment assignment (log-odds scale)
beta_t = np.array([
0.015, # tenure_months
0.10, # avg_sessions_week
0.002, # spend_last_month
0.75, # premium_user
0.30, # urban_resident: more likely to get the feature
], dtype=float)
gen = CausalDatasetGenerator(
theta=theta,
outcome_type="continuous",
sigma_y=sigma_y,
target_t_rate=target_t_rate,
seed=42,
confounder_specs=confounder_specs,
beta_y=beta_y,
beta_t=beta_t,
)
# Create dataset
causal_data = gen.to_causal_data(
n=10000,
confounders = [
"tenure_months",
"avg_sessions_week",
"spend_last_month",
"premium_user",
"urban_resident",
]
)
# Show first few rows
causal_data.df.head()
| y | t | tenure_months | avg_sessions_week | spend_last_month | premium_user | urban_resident | |
|---|---|---|---|---|---|---|---|
| 0 | 5.927714 | 1.0 | 27.656605 | 5.352554 | 72.552568 | 1.0 | 0.0 |
| 1 | 11.122008 | 1.0 | 11.520191 | 6.798247 | 188.481287 | 1.0 | 0.0 |
| 2 | 10.580393 | 1.0 | 33.005414 | 2.055459 | 51.040440 | 0.0 | 1.0 |
| 3 | 6.982844 | 1.0 | 35.286777 | 4.429404 | 166.992239 | 0.0 | 1.0 |
| 4 | 10.899381 | 0.0 | 0.587578 | 6.658307 | 179.371126 | 0.0 | 0.0 |
EDA#
from causalkit.eda import CausalEDA
eda = CausalEDA(causal_data)
---------------------------------------------------------------------------
ModuleNotFoundError Traceback (most recent call last)
Cell In[2], line 1
----> 1 from causalkit.eda import CausalEDA
2 eda = CausalEDA(causal_data)
File ~/work/CausalKit/CausalKit/causalkit/eda/__init__.py:1
----> 1 from .eda import CausalEDA, CausalDataLite
3 __all__ = ["CausalEDA", "CausalDataLite"]
File ~/work/CausalKit/CausalKit/causalkit/eda/eda.py:37
35 from sklearn.compose import ColumnTransformer
36 from sklearn.pipeline import Pipeline
---> 37 from catboost import CatBoostClassifier, CatBoostRegressor
38 import matplotlib.pyplot as plt
41 class PropensityModel:
ModuleNotFoundError: No module named 'catboost'
General dataset information#
Let’s see how outcome differ between clients who recieved the feature and didn’t
# shape of data
eda.data_shape()
{'n_rows': 10000, 'n_columns': 7}
# 1) Outcome statistics by treatment
eda.outcome_stats()
| count | mean | std | min | p10 | p25 | median | p75 | p90 | max | |
|---|---|---|---|---|---|---|---|---|---|---|
| treatment | ||||||||||
| 0.0 | 6519 | 6.011340 | 3.925893 | -9.866447 | 1.012888 | 3.426894 | 6.030504 | 8.676781 | 10.952265 | 20.770359 |
| 1.0 | 3481 | 8.553615 | 3.934260 | -5.507710 | 3.525225 | 5.909860 | 8.566860 | 11.216054 | 13.602967 | 21.377687 |
# 2) Outcome distribution by treatment (hist + boxplot)
fig1, fig2 = eda.outcome_plots()
plt.show()
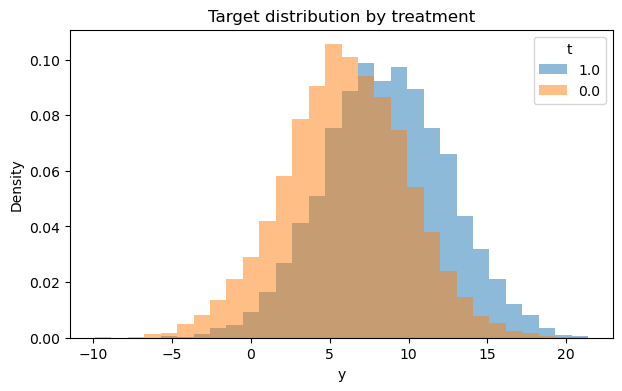
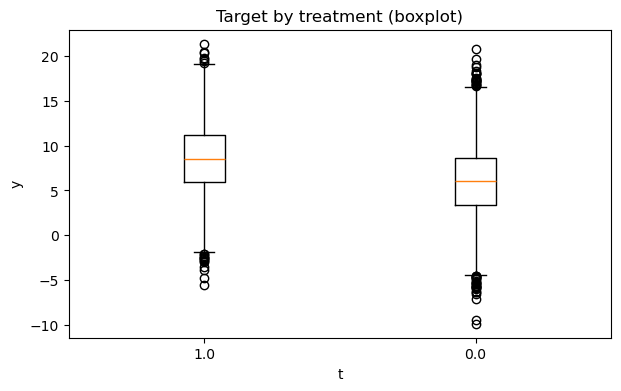
Propensity#
Now let’s examine how propensity score differ treatments
# Shows means of confounders for control/treated groups, absolute differences, and SMD values
confounders_balance_df = eda.confounders_means()
display(confounders_balance_df)
| mean_t_0 | mean_t_1 | abs_diff | smd | |
|---|---|---|---|---|
| confounders | ||||
| premium_user | 0.198343 | 0.347889 | 0.149545 | 0.340423 |
| avg_sessions_week | 4.905841 | 5.293265 | 0.387423 | 0.193959 |
| tenure_months | 23.170108 | 25.200827 | 2.030718 | 0.168676 |
| urban_resident | 0.578003 | 0.643781 | 0.065778 | 0.135209 |
| spend_last_month | 97.801230 | 105.271930 | 7.470700 | 0.129267 |
# Propensity model fit
ps_model = eda.fit_propensity()
# ROC AUC - shows how predictable treatment is from confounders
roc_auc_score = ps_model.roc_auc
print("ROC AUC from PropensityModel:", round(roc_auc_score, 4))
ROC AUC from PropensityModel: 0.6053
# Positivity check - assess overlap between treatment groups
positivity_result = ps_model.positivity_check()
print("Positivity check from PropensityModel:", positivity_result)
Positivity check from PropensityModel: {'bounds': (0.05, 0.95), 'share_below': 0.0008, 'share_above': 0.0, 'flag': False}
# SHAP values - feature importance for treatment assignment from confounders
shap_values_df = ps_model.shap
display(shap_values_df)
| feature | shap_mean | |
|---|---|---|
| 0 | spend_last_month | -0.000322 |
| 1 | tenure_months | 0.000140 |
| 2 | avg_sessions_week | 0.000122 |
| 3 | premium_user | 0.000052 |
| 4 | urban_resident | 0.000008 |
# Propensity score overlap graph
ps_model.ps_graph()
plt.show()
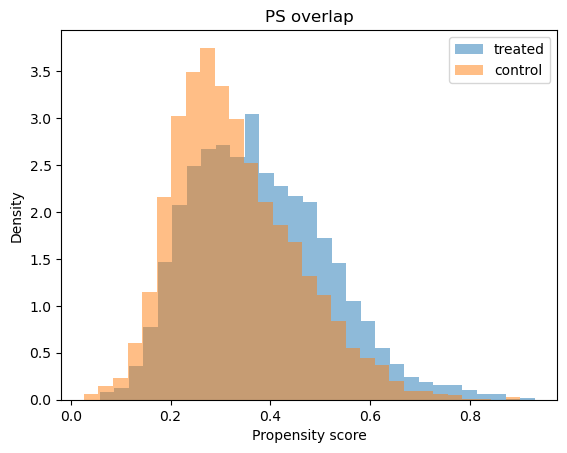
Outcome regression#
Let’s analyze how confounders predict outcome
# Outcome model fit
outcome_model = eda.outcome_fit()
# RMSE and MAE of regression model
print(outcome_model.scores)
{'rmse': 3.6896558290948964, 'mae': 2.94099663606919}
# 2) SHAP values - feature importance for outcome prediction from confounders
shap_outcome_df = outcome_model.shap
display(shap_outcome_df)
| feature | shap_mean | |
|---|---|---|
| 0 | spend_last_month | 0.002252 |
| 1 | tenure_months | -0.001747 |
| 2 | avg_sessions_week | -0.000951 |
| 3 | premium_user | 0.000729 |
| 4 | urban_resident | -0.000282 |
Inference#
Now time to estimate ATE with Double Machine Learning
from causalkit.inference.ate import dml_ate
# Estimate Average Treatment Effect (ATE)
ate_result = dml_ate(causal_data, n_folds=3, confidence_level=0.95)
ate_result
{'coefficient': 1.7547754158492368,
'std_error': 0.08120639822820347,
'p_value': 1.4835396169728733e-103,
'confidence_interval': (1.5956138000077407, 1.913937031690733),
'model': <doubleml.irm.irm.DoubleMLIRM at 0x157b9fed0>}
True theta in our data generating proccess was 1.8
Refutation#
Placebo#
# Import refutation utilities
from causalkit.refutation import (
refute_placebo_outcome,
refute_placebo_treatment,
refute_subset,
refute_irm_orthogonality,
sensitivity_analysis,
sensitivity_analysis_set
)
Replacing outcome with dummy random variable must broke our model and effect will be near zero
# Replacing an outcome with placebo
ate_placebo_outcome = refute_placebo_outcome(
dml_ate,
causal_data,
random_state=42
)
print(ate_placebo_outcome)
{'theta': 0.0010896926888271316, 'p_value': 0.863664499475262}
Replacing treatment with dummy random variable must broke our model and effect will be near zero
# Replacing treatment with placebo
ate_placebo_treatment = refute_placebo_treatment(
dml_ate,
causal_data,
random_state=42
)
print(ate_placebo_treatment)
{'theta': 0.05021278313460212, 'p_value': 0.5290821466624945}
Let’s chanllege our dataset and romove random parts. Theta shoul be near estimated
# Inference on subsets
subset_fractions = [0.3, 0.5]
ate_subset_results = []
for fraction in subset_fractions:
subset_result = refute_subset(
dml_ate,
causal_data,
fraction=fraction,
random_state=42
)
ate_subset_results.append(subset_result)
print(f" With {fraction*100:.0f}% subset: theta = {subset_result['theta']:.4f}, p_value = {subset_result['p_value']:.4f}")
With 30% subset: theta = 1.5725, p_value = 0.0000
With 50% subset: theta = 1.8468, p_value = 0.0000
Orthogonality#
Orthogonality tests show us “Does we correct specify the model”
ate_ortho_check = refute_irm_orthogonality(dml_ate, causal_data)
# 1. Out-of-sample moment check
print("\n--- 1. Out-of-Sample Moment Check ---")
oos_test = ate_ortho_check['oos_moment_test']
print(f"T-statistic: {oos_test['tstat']:.4f}")
print(f"P-value: {oos_test['pvalue']:.4f}")
print(f"Interpretation: {oos_test['interpretation']}")
print("\nFold-wise results:")
display(oos_test['fold_results'])
--- 1. Out-of-Sample Moment Check ---
T-statistic: 0.3617
P-value: 0.7176
Interpretation: Should be ≈ 0 if moment condition holds
Fold-wise results:
| fold | n | psi_mean | psi_var | |
|---|---|---|---|---|
| 0 | 0 | 2000 | 0.142070 | 64.367381 |
| 1 | 1 | 2000 | 0.114837 | 71.748247 |
| 2 | 2 | 2000 | 0.106751 | 65.548761 |
| 3 | 3 | 2000 | 0.091285 | 62.569903 |
| 4 | 4 | 2000 | -0.307471 | 68.208318 |
psi_mean is near zero on every fold, so the test is successful
# 2. Orthogonality derivatives
print("\n--- 2. Orthogonality (Gateaux Derivative) Tests ---")
ortho_derivs = ate_ortho_check['orthogonality_derivatives']
print(f"Interpretation: {ortho_derivs['interpretation']}")
print("\nFull sample derivatives:")
display(ortho_derivs['full_sample'])
print("\nTrimmed sample derivatives:")
display(ortho_derivs['trimmed_sample'])
if len(ortho_derivs['problematic_full']) > 0:
print("\n⚠ PROBLEMATIC derivatives (full sample):")
display(ortho_derivs['problematic_full'])
else:
print("\n✓ No problematic derivatives in full sample")
if len(ortho_derivs['problematic_trimmed']) > 0:
print("\n⚠ PROBLEMATIC derivatives (trimmed sample):")
display(ortho_derivs['problematic_trimmed'])
else:
print("\n✓ No problematic derivatives in trimmed sample")
--- 2. Orthogonality (Gateaux Derivative) Tests ---
Interpretation: Large |t-stats| (>2) indicate calibration issues
Full sample derivatives:
| basis | d_m1 | se_m1 | t_m1 | d_m0 | se_m0 | t_m0 | d_g | se_g | t_g | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | -0.027778 | 0.015464 | -1.796300 | 0.009315 | 0.007849 | 1.186833 | -0.103516 | 0.296654 | -0.348945 |
| 1 | 1 | 0.005088 | 0.016336 | 0.311467 | 0.003259 | 0.008088 | 0.402935 | 0.137892 | 0.322104 | 0.428097 |
| 2 | 2 | 0.003129 | 0.016049 | 0.194959 | 0.000922 | 0.008162 | 0.112934 | 0.028325 | 0.385094 | 0.073553 |
| 3 | 3 | -0.001588 | 0.014938 | -0.106328 | -0.000145 | 0.008205 | -0.017732 | 0.021827 | 0.282692 | 0.077210 |
| 4 | 4 | -0.000312 | 0.012767 | -0.024401 | 0.002863 | 0.009497 | 0.301462 | -0.019862 | 0.231095 | -0.085947 |
| 5 | 5 | 0.000451 | 0.015937 | 0.028314 | -0.001855 | 0.007660 | -0.242168 | 0.138553 | 0.316203 | 0.438177 |
Trimmed sample derivatives:
| basis | d_m1 | se_m1 | t_m1 | d_m0 | se_m0 | t_m0 | d_g | se_g | t_g | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | -0.027778 | 0.015464 | -1.796300 | 0.009315 | 0.007849 | 1.186833 | -0.103516 | 0.296654 | -0.348945 |
| 1 | 1 | 0.005088 | 0.016336 | 0.311467 | 0.003259 | 0.008088 | 0.402935 | 0.137892 | 0.322104 | 0.428097 |
| 2 | 2 | 0.003129 | 0.016049 | 0.194959 | 0.000922 | 0.008162 | 0.112934 | 0.028325 | 0.385094 | 0.073553 |
| 3 | 3 | -0.001588 | 0.014938 | -0.106328 | -0.000145 | 0.008205 | -0.017732 | 0.021827 | 0.282692 | 0.077210 |
| 4 | 4 | -0.000312 | 0.012767 | -0.024401 | 0.002863 | 0.009497 | 0.301462 | -0.019862 | 0.231095 | -0.085947 |
| 5 | 5 | 0.000451 | 0.015937 | 0.028314 | -0.001855 | 0.007660 | -0.242168 | 0.138553 | 0.316203 | 0.438177 |
✓ No problematic derivatives in full sample
✓ No problematic derivatives in trimmed sample
t_m1, t_m0, t_g should be above 2. Even after trimming our data t_m1, t_m0, t_g above 2 across all confounders
# 3. Influence diagnostics
print("\n--- 3. Influence Diagnostics ---")
influence = ate_ortho_check['influence_diagnostics']
print(f"Interpretation: {influence['interpretation']}")
print("\nFull sample influence metrics:")
print(f" Plugin SE: {influence['full_sample']['se_plugin']:.4f}")
print(f" Kurtosis: {influence['full_sample']['kurtosis']:.2f}")
print(f" P99/Median ratio: {influence['full_sample']['p99_over_med']:.2f}")
print("\nTrimmed sample influence metrics:")
print(f" Plugin SE: {influence['trimmed_sample']['se_plugin']:.4f}")
print(f" Kurtosis: {influence['trimmed_sample']['kurtosis']:.2f}")
print(f" P99/Median ratio: {influence['trimmed_sample']['p99_over_med']:.2f}")
print("\nTop influential observations (full sample):")
display(influence['full_sample']['top_influential'])
--- 3. Influence Diagnostics ---
Interpretation: Heavy tails or extreme kurtosis suggest instability
Full sample influence metrics:
Plugin SE: 0.0815
Kurtosis: 7.64
P99/Median ratio: 6.59
Trimmed sample influence metrics:
Plugin SE: 0.0815
Kurtosis: 7.64
P99/Median ratio: 6.59
Top influential observations (full sample):
| i | psi | g | res_t | res_c | |
|---|---|---|---|---|---|
| 0 | 7299 | -78.668133 | 0.100071 | -7.970469 | -0.000000 |
| 1 | 874 | 63.471801 | 0.116169 | 8.132480 | 0.000000 |
| 2 | 9405 | 49.195863 | 0.169723 | 8.457407 | 0.000000 |
| 3 | 9116 | -46.282651 | 0.164969 | -7.989467 | -0.000000 |
| 4 | 294 | -44.627395 | 0.213603 | -9.570962 | -0.000000 |
| 5 | 7634 | 44.388611 | 0.742768 | -0.000000 | -12.113462 |
| 6 | 8409 | 44.028348 | 0.284715 | 12.696785 | 0.000000 |
| 7 | 5821 | -43.885460 | 0.153316 | -6.377281 | -0.000000 |
| 8 | 8085 | -43.798883 | 0.247829 | -11.536566 | -0.000000 |
| 9 | 731 | 43.545110 | 0.165439 | 6.820135 | 0.000000 |
# Trimming information
print("\n--- Propensity Score Trimming ---")
trim_info = ate_ortho_check['trimming_info']
print(f"Trimming bounds: {trim_info['bounds']}")
print(f"Observations trimmed: {trim_info['n_trimmed']} ({trim_info['pct_trimmed']:.1f}%)")
--- Propensity Score Trimming ---
Trimming bounds: (0.02, 0.98)
Observations trimmed: 0 (0.0%)
In this test we analyze distribution of theta before trimming and after. There is no big difference. So test passed
# Diagnostic conditions breakdown
print("\n--- Diagnostic Conditions Assessment ---")
conditions = ate_ortho_check['diagnostic_conditions']
print("Individual condition checks:")
for condition, passed in conditions.items():
status = "✓ PASS" if passed else "✗ FAIL"
print(f" {condition}: {status}")
print(f"\nOverall: {ate_ortho_check['overall_assessment']}")
--- Diagnostic Conditions Assessment ---
Individual condition checks:
oos_moment_ok: ✓ PASS
derivs_full_ok: ✓ PASS
derivs_trim_ok: ✓ PASS
se_reasonable: ✓ PASS
no_extreme_influence: ✓ PASS
trimming_reasonable: ✓ PASS
Overall: PASS: Strong evidence for orthogonality
Sensitivity analysis#
Let’s analyze how unobserved confounder could look
bench_sets = causal_data.confounders
res = sensitivity_analysis_set(
ate_result,
benchmarking_set=bench_sets,
level=0.95,
null_hypothesis=0.0,
)
# Build a DataFrame with confounders as rows and the metrics as columns
summary_df = pd.DataFrame({
name: (df.loc['t'] if 't' in df.index else df.iloc[0])
for name, df in res.items()
}).T
summary_df
| cf_y | cf_d | rho | delta_theta | |
|---|---|---|---|---|
| tenure_months | 0.019343 | 0.004555 | 1.000000 | 0.125310 |
| avg_sessions_week | 0.031075 | 0.010474 | 1.000000 | 0.172854 |
| spend_last_month | 0.080634 | 0.006735 | 0.982448 | 0.177995 |
| premium_user | 0.062270 | 0.031318 | 1.000000 | 0.360557 |
| urban_resident | 0.018716 | 0.000000 | 1.000000 | 0.093394 |
It is business domain and data knowledge relative question. In this situation we will stop on the most influential confounder ‘premium_user’ and test theta when we do not observe anothere confounder with strength of ‘premium_user’
# Run sensitivity analysis on our ATE result
sensitivity_report_1 = sensitivity_analysis(
ate_result,
cf_y=0.06, # Confounding strength affecting outcome
cf_d=0.03, # Confounding strength affecting treatment
rho=1.0 # Perfect correlation between unobserved confounders
)
print(sensitivity_report_1)
================== Sensitivity Analysis ==================
------------------ Scenario ------------------
Significance Level: level=0.95
Sensitivity parameters: cf_y=0.06; cf_d=0.03, rho=1.0
------------------ Bounds with CI ------------------
CI lower theta lower theta theta upper CI upper
t 1.284916 1.418741 1.754775 2.09081 2.224353
------------------ Robustness Values ------------------
H_0 RV (%) RVa (%)
t 0.0 20.106836 18.701604
CI lower >> 0. It means test is passed