DML ATT Example#
This notebook covers scenario:
Is RCT |
Treatment |
Outcome |
EDA |
Estimands |
Refutation |
|---|---|---|---|---|---|
Observational |
Binary |
Continuous |
Yes |
ATT |
Yes |
We will estimate Average Treatment Effect on Treated (ATT) of binary treatment on continuous outcome. It shows explonatary data analysis and refutation tests
Generate data#
Example that generates observational data with a nonlinear outcome model, nonlinear treatment assignment, and a heterogeneous (nonlinear) treatment effect tau(X). This setup ensures that ATT ≠ ATE in general. It also shows how to compute the “ground-truth” ATT from the generated data.
# Nonlinear ATT data generation with heterogeneous effects
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from causalkit.data import CausalDatasetGenerator, CausalData
# Reproducibility
np.random.seed(42)
# 1) Confounders and their distributions
# These names define the column order in X for the custom functions.
confounder_specs = [
{"name": "tenure_months", "dist": "normal", "mu": 24, "sd": 12},
{"name": "avg_sessions_week", "dist": "normal", "mu": 5, "sd": 2},
{"name": "spend_last_month", "dist": "uniform", "a": 0, "b": 200},
{"name": "premium_user", "dist": "bernoulli","p": 0.25},
{"name": "urban_resident", "dist": "bernoulli","p": 0.60},
]
# Indices (for convenience inside g_y, g_t, tau)
TENURE, SESS, SPEND, PREMIUM, URBAN = range(5)
# 2) Nonlinear baseline for outcome f_y(X) = X @ beta_y + g_y(X)
# Keep a modest linear part and add meaningful nonlinearities.
beta_y = np.array([
0.03, # tenure_months
0.20, # avg_sessions_week
0.01, # spend_last_month
1.20, # premium_user
0.60, # urban_resident
], dtype=float)
def g_y(X: np.ndarray) -> np.ndarray:
# Nonlinearities and interactions in outcome baseline
tenure_years = X[:, TENURE] / 12.0
sessions = X[:, SESS]
spend = X[:, SPEND]
premium = X[:, PREMIUM]
urban = X[:, URBAN]
return (
1.2 * np.sin(2.0 * np.pi * tenure_years) # seasonal-ish tenure pattern
+ 0.02 * (sessions - 5.0) ** 2 # convex effect of sessions
+ 0.0015 * (spend - 100.0) * (sessions - 5.0) # spend × sessions interaction
+ 0.4 * premium * (sessions - 5.0) # premium × sessions interaction
+ 0.3 * urban * np.tanh((spend - 100.0) / 50.0) # nonlinear spend effect differs by urban
)
# 3) Nonlinear treatment score f_t(X) = X @ beta_t + g_t(X)
beta_t = np.array([
0.010, # tenure_months
0.12, # avg_sessions_week
0.001, # spend_last_month
0.80, # premium_user
0.25, # urban_resident
], dtype=float)
def g_t(X: np.ndarray) -> np.ndarray:
tenure_years = X[:, TENURE] / 12.0
sessions = X[:, SESS]
spend = X[:, SPEND]
premium = X[:, PREMIUM]
urban = X[:, URBAN]
# Smoothly increasing selection with spend; interactions make selection non-separable
soft_spend = 1.2 * np.tanh((spend - 80.0) / 40.0)
return (
0.6 * soft_spend
+ 0.15 * (sessions - 5.0) * (tenure_years - 2.0)
+ 0.25 * premium * (urban - 0.5)
)
# 4) Heterogeneous, nonlinear treatment effect tau(X) on the natural scale (continuous outcome)
def tau_fn(X: np.ndarray) -> np.ndarray:
tenure_years = X[:, TENURE] / 12.0
sessions = X[:, SESS]
spend = X[:, SPEND]
premium = X[:, PREMIUM]
urban = X[:, URBAN]
# Base effect + stronger effect for higher sessions and premium users,
# diminishes with tenure, mild modulation by spend and urban
tau = (
1.0
+ 0.8 * (1.0 / (1.0 + np.exp(-(sessions - 5.0)))) # sigmoid in sessions
+ 0.5 * premium
- 0.6 * np.clip(tenure_years / 5.0, 0.0, 1.0) # taper with long tenure
+ 0.2 * urban * (spend - 100.0) / 100.0
)
# Optional: keep it in a reasonable range
return np.clip(tau, 0.2, 2.5)
# 5) Noise and prevalence
sigma_y = 3.5
target_t_rate = 0.35 # enforce ~35% treated via intercept calibration
# 6) Build generator
gen = CausalDatasetGenerator(
outcome_type="continuous",
sigma_y=sigma_y,
target_t_rate=target_t_rate,
seed=42,
# Confounders
confounder_specs=confounder_specs,
# Outcome/treatment structure
beta_y=beta_y,
beta_t=beta_t,
g_y=g_y,
g_t=g_t,
# Heterogeneous effect
tau=tau_fn,
)
# 7) Generate data (full dataframe includes ground-truth columns: propensity, mu0, mu1, cate)
n = 10000
generated_df = gen.generate(n)
# Ground-truth ATT (on the natural scale): E[tau(X) | T=1] = mean CATE among the treated
true_att = float(generated_df.loc[generated_df["t"] == 1, "cate"].mean())
print(f"Ground-truth ATT from the DGP: {true_att:.3f}")
# 8) Wrap as CausalData for downstream workflows (keeps only y, t, and specified confounders)
causal_data = CausalData(
df=generated_df,
treatment="t",
outcome="y",
confounders=[
"tenure_months",
"avg_sessions_week",
"spend_last_month",
"premium_user",
"urban_resident",
],
)
# Peek at the analysis-ready view
causal_data.df.head()
Ground-truth ATT from the DGP: 1.385
| y | t | tenure_months | avg_sessions_week | spend_last_month | premium_user | urban_resident | |
|---|---|---|---|---|---|---|---|
| 0 | 3.930539 | 1.0 | 27.656605 | 5.352554 | 72.552568 | 1.0 | 0.0 |
| 1 | 5.771469 | 0.0 | 11.520191 | 6.798247 | 188.481287 | 1.0 | 0.0 |
| 2 | 6.374653 | 1.0 | 33.005414 | 2.055459 | 51.040440 | 0.0 | 1.0 |
| 3 | 2.364177 | 1.0 | 35.286777 | 4.429404 | 166.992239 | 0.0 | 1.0 |
| 4 | 8.378079 | 0.0 | 0.587578 | 6.658307 | 179.371126 | 0.0 | 0.0 |
EDA#
from causalkit.eda import CausalEDA
eda = CausalEDA(causal_data)
---------------------------------------------------------------------------
ModuleNotFoundError Traceback (most recent call last)
Cell In[2], line 1
----> 1 from causalkit.eda import CausalEDA
2 eda = CausalEDA(causal_data)
File ~/work/CausalKit/CausalKit/causalkit/eda/__init__.py:1
----> 1 from .eda import CausalEDA, CausalDataLite
3 __all__ = ["CausalEDA", "CausalDataLite"]
File ~/work/CausalKit/CausalKit/causalkit/eda/eda.py:37
35 from sklearn.compose import ColumnTransformer
36 from sklearn.pipeline import Pipeline
---> 37 from catboost import CatBoostClassifier, CatBoostRegressor
38 import matplotlib.pyplot as plt
41 class PropensityModel:
ModuleNotFoundError: No module named 'catboost'
General dataset information#
Let’s see how outcome differ between clients who recieved the feature and didn’t
# shape of data
eda.data_shape()
{'n_rows': 10000, 'n_columns': 7}
# 1) Outcome statistics by treatment
eda.outcome_stats()
| count | mean | std | min | p10 | p25 | median | p75 | p90 | max | |
|---|---|---|---|---|---|---|---|---|---|---|
| treatment | ||||||||||
| 0.0 | 6526 | 3.189869 | 3.755023 | -12.770866 | -1.572612 | 0.699795 | 3.149570 | 5.723303 | 7.935340 | 17.323664 |
| 1.0 | 3474 | 5.216191 | 3.987103 | -7.310514 | 0.147968 | 2.504771 | 5.199664 | 7.959577 | 10.348905 | 18.147842 |
# 2) Outcome distribution by treatment (hist + boxplot)
fig1, fig2 = eda.outcome_plots()
plt.show()
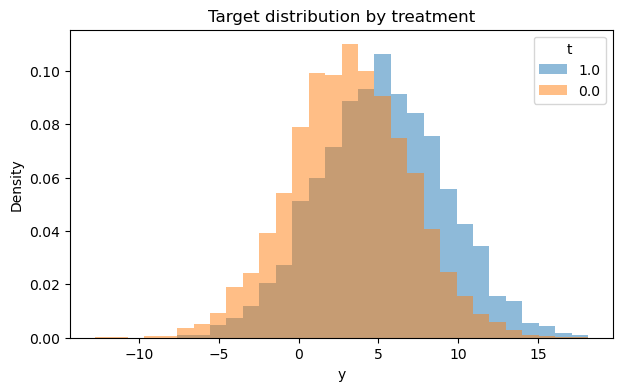
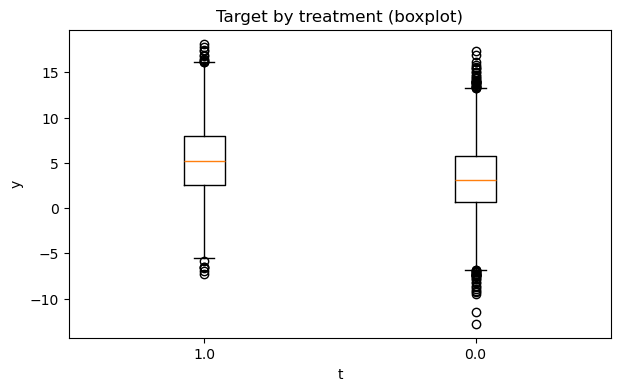
Propensity#
Now let’s examine how propensity score differ treatments
# Shows means of confounders for control/treated groups, absolute differences, and SMD values
confounders_balance_df = eda.confounders_means()
display(confounders_balance_df)
| mean_t_0 | mean_t_1 | abs_diff | smd | |
|---|---|---|---|---|
| confounders | ||||
| spend_last_month | 90.313423 | 119.353025 | 29.039602 | 0.523162 |
| premium_user | 0.195066 | 0.354347 | 0.159281 | 0.362615 |
| avg_sessions_week | 4.902830 | 5.299702 | 0.396872 | 0.198761 |
| urban_resident | 0.576463 | 0.646805 | 0.070341 | 0.144688 |
| tenure_months | 23.329078 | 24.906290 | 1.577212 | 0.130648 |
# Propensity model fit
ps_model = eda.fit_propensity()
# ROC AUC - shows how predictable treatment is from confounders
roc_auc_score = ps_model.roc_auc
print("ROC AUC from PropensityModel:", round(roc_auc_score, 4))
ROC AUC from PropensityModel: 0.6937
# Positivity check - assess overlap between treatment groups
positivity_result = ps_model.positivity_check()
print("Positivity check from PropensityModel:", positivity_result)
Positivity check from PropensityModel: {'bounds': (0.05, 0.95), 'share_below': 0.0132, 'share_above': 0.0004, 'flag': False}
# SHAP values - feature importance for treatment assignment from confounders
shap_values_df = ps_model.shap
display(shap_values_df)
| feature | shap_mean | |
|---|---|---|
| 0 | spend_last_month | -0.000735 |
| 1 | tenure_months | 0.000451 |
| 2 | urban_resident | 0.000190 |
| 3 | premium_user | 0.000124 |
| 4 | avg_sessions_week | -0.000029 |
# Propensity score overlap graph
ps_model.ps_graph()
plt.show()
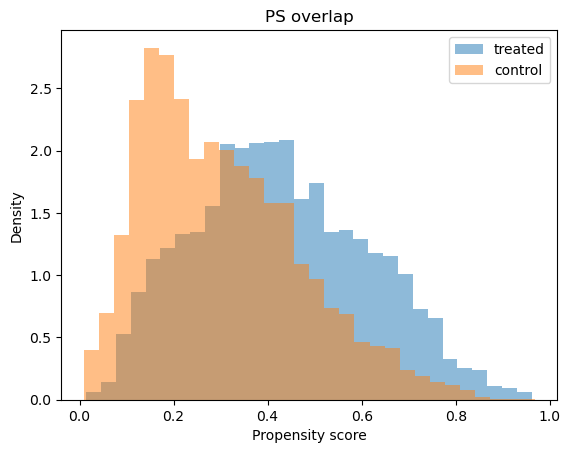
Outcome regression#
Let’s analyze how confounders predict outcome
# Outcome model fit
outcome_model = eda.outcome_fit()
# RMSE and MAE of regression model
print(outcome_model.scores)
{'rmse': 3.6452253851701975, 'mae': 2.905800097741897}
# 2) SHAP values - feature importance for outcome prediction from confounders
shap_outcome_df = outcome_model.shap
display(shap_outcome_df)
| feature | shap_mean | |
|---|---|---|
| 0 | premium_user | 0.001378 |
| 1 | avg_sessions_week | -0.001012 |
| 2 | spend_last_month | -0.000987 |
| 3 | urban_resident | 0.000543 |
| 4 | tenure_months | 0.000078 |
Inference#
Now time to estimate ATE with Double Machine Learning
from causalkit.inference.att import dml_att
# Estimate Average Treatment Effect (ATT)
att_result = dml_att(causal_data, n_folds=3, confidence_level=0.95)
att_result
{'coefficient': 1.3111886779440418,
'std_error': 0.09481720872967514,
'p_value': 1.7133685893213766e-43,
'confidence_interval': (1.1253503637192617, 1.497026992168822),
'model': <doubleml.irm.irm.DoubleMLIRM at 0x163ba74d0>}
Real ATT is 1.385
Refutation#
# Import refutation utilities
from causalkit.refutation import (
refute_placebo_outcome,
refute_placebo_treatment,
refute_subset,
refute_irm_orthogonality,
sensitivity_analysis,
sensitivity_analysis_set
)
Placebo#
Replacing outcome with dummy random variable must broke our model and effect will be near zero
# Replacing an outcome with placebo
att_placebo_outcome = refute_placebo_outcome(
dml_att,
causal_data,
random_state=42
)
print(att_placebo_outcome)
{'theta': 0.0024891263554991036, 'p_value': 0.49268997429089034}
Replacing treatment with dummy random variable must broke our model and effect will be near zero
# Replacing treatment with placebo
att_placebo_treatment = refute_placebo_treatment(
dml_att,
causal_data,
random_state=42
)
print(att_placebo_treatment)
{'theta': -0.004944799398107784, 'p_value': 0.9497270383821083}
Let’s chanllege our dataset and romove random parts. Theta shoul be near estimated
# Inference on subsets
subset_fractions = [0.3, 0.5]
att_subset_results = []
for fraction in subset_fractions:
subset_result = refute_subset(
dml_att,
causal_data,
fraction=fraction,
random_state=42
)
att_subset_results.append(subset_result)
print(f" With {fraction*100:.0f}% subset: theta = {subset_result['theta']:.4f}, p_value = {subset_result['p_value']:.4f}")
With 30% subset: theta = 1.1546, p_value = 0.0000
With 50% subset: theta = 1.3733, p_value = 0.0000
Orthogonality#
Orthogonality tests validate whether our ATT estimator is properly specified. We will inspect:
Out-of-sample (OOS) moment check;
ATT-specific orthogonality derivatives (m0 and g only);
Influence diagnostics;
ATT overlap diagnostics and trim-sensitivity near m→1 (controls);
Trimming info and overall diagnostic conditions.
att_ortho_check = refute_irm_orthogonality(dml_att, causal_data, target='ATT')
1. Out-of-sample moment check#
print("\n--- 1. Out-of-Sample Moment Check ---")
oos_test = att_ortho_check['oos_moment_test']
print(f"T-statistic: {oos_test['tstat']:.4f}")
print(f"P-value: {oos_test['pvalue']:.4f}")
print(f"Interpretation: {oos_test['interpretation']}")
print("\nFold-wise results:")
display(oos_test['fold_results'])
--- 1. Out-of-Sample Moment Check ---
T-statistic: -0.2644
P-value: 0.7915
Interpretation: Should be ≈ 0 if moment condition holds
Fold-wise results:
| fold | n | psi_mean | psi_var | |
|---|---|---|---|---|
| 0 | 0 | 2000 | 0.329863 | 83.664056 |
| 1 | 1 | 2000 | -0.573543 | 177.284891 |
| 2 | 2 | 2000 | 0.275107 | 89.104143 |
| 3 | 3 | 2000 | 0.177037 | 66.171971 |
| 4 | 4 | 2000 | -0.339714 | 76.546856 |
psi_mean is expected to be near zero on every fold if the moment condition holds.
2. Orthogonality derivatives (ATT)#
print("\n--- 2. Orthogonality (Gateaux Derivative) Tests — ATT ---")
ortho_derivs = att_ortho_check['orthogonality_derivatives']
print(f"Interpretation: {ortho_derivs['interpretation']}")
print("\nFull sample derivatives:")
display(ortho_derivs['full_sample'])
print("\nTrimmed sample derivatives:")
display(ortho_derivs['trimmed_sample'])
if len(ortho_derivs['problematic_full']) > 0:
print("\n\u26A0 PROBLEMATIC derivatives (full sample):")
display(ortho_derivs['problematic_full'])
else:
print("\n\u2713 No problematic derivatives in full sample")
if len(ortho_derivs['problematic_trimmed']) > 0:
print("\n\u26A0 PROBLEMATIC derivatives (trimmed sample):")
display(ortho_derivs['problematic_trimmed'])
else:
print("\n\u2713 No problematic derivatives in trimmed sample")
--- 2. Orthogonality (Gateaux Derivative) Tests — ATT ---
Interpretation: ATT: check m0 & g only; large |t| (>2) => calibration issues
Full sample derivatives:
| basis | d_m1 | se_m1 | t_m1 | d_m0 | se_m0 | t_m0 | d_g | se_g | t_g | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 0.0 | 0.0 | 0.0 | 0.029850 | 0.025167 | 1.186063 | -1.009071 | 1.075327 | -0.938385 |
| 1 | 1 | 0.0 | 0.0 | 0.0 | 0.011241 | 0.029021 | 0.387342 | -2.351915 | 2.324239 | -1.011908 |
| 2 | 2 | 0.0 | 0.0 | 0.0 | 0.005547 | 0.028698 | 0.193305 | -1.938678 | 2.048945 | -0.946184 |
| 3 | 3 | 0.0 | 0.0 | 0.0 | 0.022577 | 0.025612 | 0.881506 | -1.069048 | 1.040382 | -1.027554 |
| 4 | 4 | 0.0 | 0.0 | 0.0 | 0.000924 | 0.031841 | 0.029025 | -2.086775 | 1.827563 | -1.141835 |
| 5 | 5 | 0.0 | 0.0 | 0.0 | -0.002677 | 0.024129 | -0.110944 | -0.829083 | 0.892210 | -0.929247 |
Trimmed sample derivatives:
| basis | d_m1 | se_m1 | t_m1 | d_m0 | se_m0 | t_m0 | d_g | se_g | t_g | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 0.0 | 0.0 | 0.0 | 0.029844 | 0.025167 | 1.185838 | -1.009999 | 1.075327 | -0.939249 |
| 1 | 1 | 0.0 | 0.0 | 0.0 | 0.011255 | 0.029021 | 0.387832 | -2.349584 | 2.324238 | -1.010905 |
| 2 | 2 | 0.0 | 0.0 | 0.0 | 0.005538 | 0.028698 | 0.192966 | -1.940275 | 2.048944 | -0.946963 |
| 3 | 3 | 0.0 | 0.0 | 0.0 | 0.022581 | 0.025612 | 0.881691 | -1.068271 | 1.040382 | -1.026807 |
| 4 | 4 | 0.0 | 0.0 | 0.0 | 0.000927 | 0.031841 | 0.029127 | -2.086239 | 1.827563 | -1.141541 |
| 5 | 5 | 0.0 | 0.0 | 0.0 | -0.002670 | 0.024129 | -0.110656 | -0.827944 | 0.892210 | -0.927971 |
✓ No problematic derivatives in full sample
✓ No problematic derivatives in trimmed sample
For ATT, focus on t-statistics for m0 and g. Large absolute t-values (|t| > 2) suggest calibration issues.
3. Influence diagnostics#
print("\n--- 3. Influence Diagnostics ---")
influence = att_ortho_check['influence_diagnostics']
print(f"Interpretation: {influence['interpretation']}")
print("\nFull sample influence metrics:")
print(f" Plugin SE: {influence['full_sample']['se_plugin']:.4f}")
print(f" Kurtosis: {influence['full_sample']['kurtosis']:.2f}")
print(f" P99/Median ratio: {influence['full_sample']['p99_over_med']:.2f}")
print("\nTrimmed sample influence metrics:")
print(f" Plugin SE: {influence['trimmed_sample']['se_plugin']:.4f}")
print(f" Kurtosis: {influence['trimmed_sample']['kurtosis']:.2f}")
print(f" P99/Median ratio: {influence['trimmed_sample']['p99_over_med']:.2f}")
print("\nTop influential observations (full sample):")
display(influence['full_sample']['top_influential'])
--- 3. Influence Diagnostics ---
Interpretation: Heavy tails or extreme kurtosis suggest instability
Full sample influence metrics:
Plugin SE: 0.0993
Kurtosis: 372.97
P99/Median ratio: 8.23
Trimmed sample influence metrics:
Plugin SE: 0.0993
Kurtosis: 372.66
P99/Median ratio: 8.20
Top influential observations (full sample):
| i | psi | g | res_t | res_c | |
|---|---|---|---|---|---|
| 0 | 5169 | -433.731443 | 0.955891 | 0.0 | 6.953028 |
| 1 | 9369 | -115.655411 | 0.768881 | 0.0 | 12.077354 |
| 2 | 5325 | -92.729291 | 0.760111 | 0.0 | 10.166691 |
| 3 | 9615 | -92.694582 | 0.829842 | 0.0 | 6.602995 |
| 4 | 4002 | -80.529155 | 0.768305 | 0.0 | 8.436566 |
| 5 | 5990 | 77.598309 | 0.774365 | -0.0 | -7.854934 |
| 6 | 8342 | -67.190692 | 0.776890 | 0.0 | 6.703443 |
| 7 | 313 | -63.173514 | 0.777839 | 0.0 | 6.268191 |
| 8 | 7152 | -61.702928 | 0.802739 | 0.0 | 5.267487 |
| 9 | 2958 | -59.609550 | 0.721210 | 0.0 | 8.005001 |
4. ATT overlap diagnostics and trim-sensitivity#
print("\n--- ATT Overlap Diagnostics ---")
overlap = att_ortho_check['overlap_diagnostics']
if overlap is not None:
display(overlap)
else:
print("Overlap diagnostics not available.")
print("\n--- ATT Trim-Sensitivity Curve ---")
robust = att_ortho_check['robustness']
trim_curve = robust.get('trim_curve', None) if robust is not None else None
if trim_curve is not None:
display(trim_curve)
else:
print("Trim-sensitivity curve not available.")
--- ATT Overlap Diagnostics ---
| threshold | pct_controls_with_g_ge_thr | pct_treated_with_g_le_1_minus_thr | |
|---|---|---|---|
| 0 | 0.95 | 0.015323 | 0.028785 |
| 1 | 0.97 | 0.000000 | 0.000000 |
| 2 | 0.98 | 0.000000 | 0.000000 |
| 3 | 0.99 | 0.000000 | 0.000000 |
--- ATT Trim-Sensitivity Curve ---
| trim_threshold | n | pct_dropped | theta | se | |
|---|---|---|---|---|---|
| 0 | 0.900000 | 9999 | 0.01 | 1.350662 | 0.090084 |
| 1 | 0.908636 | 9999 | 0.01 | 1.331246 | 0.091142 |
| 2 | 0.917273 | 9999 | 0.01 | 1.387317 | 0.092264 |
| 3 | 0.925909 | 9999 | 0.01 | 1.347702 | 0.090716 |
| 4 | 0.934545 | 9999 | 0.01 | 1.359888 | 0.091188 |
| 5 | 0.943182 | 9999 | 0.01 | 1.356327 | 0.090682 |
| 6 | 0.951818 | 9999 | 0.01 | 1.352705 | 0.091355 |
| 7 | 0.960455 | 10000 | 0.00 | 1.292609 | 0.096413 |
| 8 | 0.969091 | 10000 | 0.00 | 1.328536 | 0.094768 |
| 9 | 0.977727 | 10000 | 0.00 | 1.351585 | 0.092971 |
| 10 | 0.986364 | 10000 | 0.00 | 1.346071 | 0.095060 |
| 11 | 0.995000 | 10000 | 0.00 | 1.359726 | 0.092266 |
5. Propensity score trimming#
print("\n--- Propensity Score Trimming ---")
trim_info = att_ortho_check['trimming_info']
print(f"Trimming bounds: {trim_info['bounds']}")
print(f"Observations trimmed: {trim_info['n_trimmed']} ({trim_info['pct_trimmed']:.1f}%)")
--- Propensity Score Trimming ---
Trimming bounds: (0.02, 0.98)
Observations trimmed: 1 (0.0%)
6. Diagnostic conditions breakdown#
print("\n--- Diagnostic Conditions Assessment ---")
conditions = att_ortho_check['diagnostic_conditions']
print("Individual condition checks:")
for condition, passed in conditions.items():
status = "\u2713 PASS" if passed else "\u2717 FAIL"
print(f" {condition}: {status}")
print(f"\nOverall: {att_ortho_check['overall_assessment']}")
--- Diagnostic Conditions Assessment ---
Individual condition checks:
oos_moment_ok: ✓ PASS
derivs_full_ok: ✓ PASS
derivs_trim_ok: ✓ PASS
se_reasonable: ✓ PASS
no_extreme_influence: ✓ PASS
trimming_reasonable: ✓ PASS
Overall: PASS: Strong evidence for orthogonality
Sensitivity analysis#
Let’s analyze how unobserved confounder could look
bench_sets = causal_data.confounders
res = sensitivity_analysis_set(
att_result,
benchmarking_set=bench_sets,
level=0.95,
null_hypothesis=0.0,
)
# Build a DataFrame with confounders as rows and the metrics as columns
summary_df = pd.DataFrame({
name: (df.loc['t'] if 't' in df.index else df.iloc[0])
for name, df in res.items()
}).T
summary_df
| cf_y | cf_d | rho | delta_theta | |
|---|---|---|---|---|
| tenure_months | 0.052112 | 0.004799 | 0.534682 | 0.069932 |
| avg_sessions_week | 0.033761 | 0.017667 | 0.828866 | 0.166359 |
| spend_last_month | 0.023657 | 0.081000 | 1.000000 | 0.385810 |
| premium_user | 0.039786 | 0.047103 | 0.646352 | 0.226695 |
| urban_resident | 0.000000 | 0.011797 | 1.000000 | 0.023253 |
It is business domain and data knowledge relative question. In this situation we will stop on the most influential confounder ‘spend_last_month’ and test theta when we do not observe another confounder with strength of ‘premium_user’
# Run sensitivity analysis on our ATT result
sensitivity_report_1 = sensitivity_analysis(
att_result,
cf_y=0.02, # Confounding strength affecting outcome
cf_d=0.08, # Confounding strength affecting treatment
rho=1.0 # Perfect correlation between unobserved confounders
)
print(sensitivity_report_1)
================== Sensitivity Analysis ==================
------------------ Scenario ------------------
Significance Level: level=0.95
Sensitivity parameters: cf_y=0.02; cf_d=0.08, rho=1.0
------------------ Bounds with CI ------------------
CI lower theta lower theta theta upper CI upper
t 0.804583 0.965456 1.311189 1.656921 1.809618
------------------ Robustness Values ------------------
H_0 RV (%) RVa (%)
t 0.0 14.614512 12.772404
CI lower >> 0. It means test is passed